Superfamily Affair: Refining the Beetle Tree
Every beetle fan knows the story: “Darwin was peeling bark and collecting beetles. He had one beetle in each hand already, and he saw a third that was exciting to him, so he popped one of the ones in his hands into his mouth.” Curator and Schlinger Chair of Entomology Matthew L. Gimmel, Ph.D., told me this story the day I met him. The beetle Darwin put in his mouth to free up one hand “turned out to be one with really foul-tasting chemical defenses, and it released in his mouth. So he, of course, spit it out. He ended up losing all three beetles in the process.”
It’s a tale told so many times that Google knows when you’re about to ask for it, and there’s a reason for that.
Not only is it a wonderful illustration of Darwin’s personality, the story illuminates a larger trend among coleopterists (those who study beetles, a.k.a. Coleoptera). They are unusually passionate about their field of study. “Beetle collectors get very obsessive,” admits Dr. Gimmel. While I haven’t yet seen Gimmel put any beetles in his mouth, I can’t shake the feeling that he’d probably be up for it. After all, it seems like a very short step from sucking up bugs with an aspirator—a widely used collection technique among entomologists—to attempting this dubious short-term storage method.

Gimmel demonstrates how to beat a bush and suck up falling bugs to Leadership Circles of Giving members during a behind-the-scenes Science Salon in the Department of Invertebrate Zoology.
Gimmel grew up in Oklahoma and remembers being passionate about natural history from a young age. His father took him on fossil-hunting field trips, and his interests cycled through phases that encompassed rocks and minerals, trees, birds, and reptiles and amphibians. He particularly enjoyed the latter because they could be kept alive at home for close-up observation. However, the young naturalist lost interest as he ran out of local species to add to his collection. In his mid-teens, he discovered beetles, being bitten by the beetle bug* in the form of a Peterson field guide to North American beetles. The guide was more comprehensive than the disappointingly sketchy field guide to insects in general, and it showed a far greater diversity of species than that seen among reptiles and amphibians. “That’s what really got me going,” Gimmel recalls. “The author, Richard White, hand-illustrated everything in that guide, with hundreds of illustrations.” Inspired by the tiny dots illustrating the actual sizes of miniscule beetles, he got a microscope for Christmas, and started following White’s instructions for assembling a collection.

Gimmel still has specimens from these early days in his private beetle collection. You’d never know by looking that the Strategus mormon (largest beetle in photo) was captured in the parking lot of an Oklahoma pizza parlor, or that the beautiful Phanaeus vindex (iridescent beetle in foreground) was lured by a pile of dog poop in Gimmel’s backyard.
“We had a suburban backyard, nothing particularly special…but there were lots and lots of beetles, and I could find new ones every day.” Gimmel describes what made this so special, using the glowing tones people normally reserve to describe their favorite desserts: “I could collect them and mount them and keep them in nice little rows in my box, and preserve them forever and look at them under the microscope whenever I wanted to, and they’d basically look how they did when I first collected them. With beetles, it was endless fun.”

Paracotalpa ursina in SBMNH collection. Now isn’t that satisfying?
“Endless” is right. The diversity of invertebrate life in general seems to provide infinite work for taxonomists, and beetles account for a significant portion of this eternal game of catch-up, as animals go extinct (and diverge into new species) faster than we can keep track of them. One consequence of this is the likelihood that whatever you are planning to do with beetles will turn into more than you bargained for, as it did for Darwin. That was certainly the case with a major project Dr. Gimmel just saw come to fruition in the form of a published monograph: a thorough study of the genetic relationships in the superfamily Cleroidea, a group of beetles containing over 10,000 identified species.†
The term superfamily makes me think of the Jackson 5, but it refers to something different. (Skip this paragraph if you already know what a superfamily is.) It’s the taxonomic level used to classify a group of organisms that are related at a certain level. Most of us are familiar with the classifying levels of species and genus, and if we stretch our minds back to high school biology, we may recall that above genus is family, and above that, in ascending sequence, are order, class, phylum, and kingdom. If your biology teacher acknowledged the incredible success of Bacteria and Archaea, you learned the highest level, known as domain. At the domain level, we distinguish those two groups from Eukarya (the domain including the large-scale life forms who so often imagine the world revolves around them). So a superfamily is a group of organisms more closely related than those in an order, but more wide-ranging than a family. The mnemonic “dirty King Philip calls out for good soup” (which can be freely adjusted to something more memorable as needed) helps some people remember the sequence of levels.
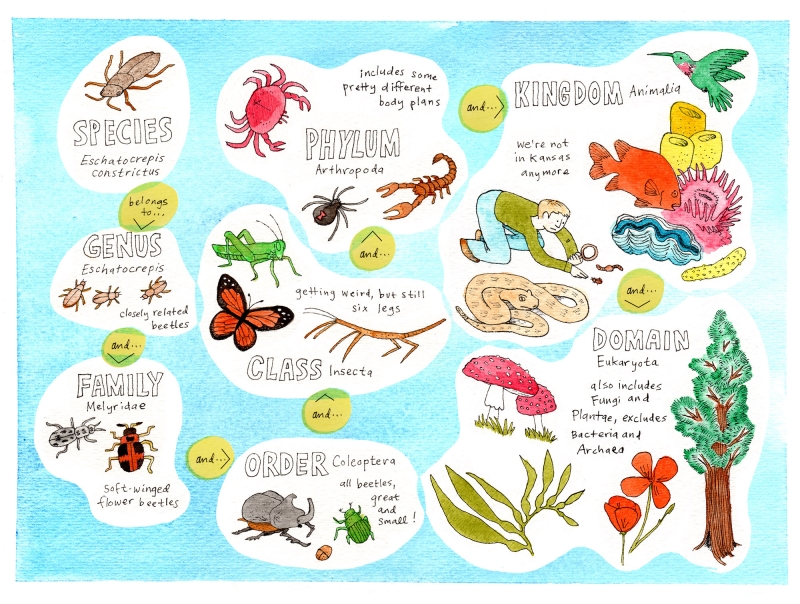
An example of how a given species also belongs to a cascade of higher-level taxonomic groups including increasingly distant relatives
At 10,000 and growing (it’s estimated that the superfamily actually contains 20-30,000 species, but we just haven’t discovered them yet), Cleroidea is merely the tenth largest superfamily among beetles. The group attracted Gimmel’s attention when he was a humble postdoc starting in 2013, in part because the superfamily contained families that had received uneven levels of attention by scholars. His postdoc supervisor, Dr. Milada Bocakova at Palacky University in the Czech Republic (a coauthor on the Cleroidea paper), had already traced the tree of evolutionary relationships in the family Melyridae. Gimmel’s fellow postdoc, Nicole L. Gunter (another coauthor), had sorted out family Cleridae. Gimmel wanted to do the same for the family Trogossitidae, which appeared to be in serious need of tidying up. How can you tell when your taxonomy needs tidying? Here’s a clue: organisms that look radically different—and have different lifestyles—are lumped together in ways that don’t make sense.

Left to right: Fungus-eating, microscopic Rentonium bicolor and good-sized predatory beetle Egolia variegata, once classified together in family Trogossitidae, but no more, thanks to Gimmel et al.’s analysis of their DNA. Photos by Matthew and Lucie Gimmel, respectively
Ways that don’t make sense to experts, I hasten to add. Some things make no sense in appearance to outsiders, but are evident to those concerned. Why would a grown man travel to New Zealand, find a rotting log, and bring it to his mentor’s house, where the two of them would sit in the basement, breaking open nodules of yellow fungus in search of grubs? To collect and study the previously-undescribed larvae of Rentonium bicolor, of course! It was in these circumstances that Gimmel developed his thirst for knowledge of the true relationships among the beetles taxonomists had assigned to family Trogossitidae.

The fungus-covered log that really tied the room together
As adults, the so-called trogossitids whose larvae Gimmel and Dr. Richard Leschen (the final coauthor) were studying bore little resemblance to others in the Trogossitidae family. It seemed to Gimmel that they could not possibly be closely related. “This kinda got me jazzed about where these might be in the tree of beetles in general,” Gimmel recalls. “We were pretty convinced they’re in this Cleroidea superfamily, and also pretty convinced that they can’t be close with trogossitids. But nobody had any other ideas where to place them.” Gimmel and Leschen were looking at the larvae in the hopes that they might see characteristics which would turn out to be family resemblances.
Most research focuses on the adult stage of an insect’s life, but as Gimmel notes, “Sometimes when you look at beetle larvae, you get more insights. Adults are just one set of the expression of their genes and evolution, right? You have to look at larvae, too. Eggs and pupae, also, but almost nobody looks at those things.” When I point out that ignoring eggs, larvae, and pupae is blatantly ageist, he agrees. “Yes, exactly. Stage-ist, right?”
In the end, even studying the external characteristics of the larval stage didn’t answer the questions about family relationships. So Gimmel conceived an ambitious project to analyze their genes instead. Why not examine the DNA of family Trogossitidae, factor in the data from Bocakova and Gunter’s studies on Melyridae and Cleridae, bring in some more species to represent the other families in Cleroidea, and make it a superfamily affair? Such a project would make excellent use of existing information and extend it to illuminate a much larger group. “I thought that it would be fairly limited, as you do at the beginning of a project,” sighs Gimmel, “but it turned out to be this massive dataset in the end. I just kept gathering material.”
While a Ph.D. student, Gimmel had sequenced a single gene for a dozen species, “just to get [his] feet wet,” but he wanted more experience doing molecular (DNA-based) research. Embarking on a project that would eventually involve examining 395 different species—looking at four genes of each—was a big jump. Furthermore, he wanted the study to be wide-ranging and representative of diversity across the superfamily, which required carefully selecting and securing specimens from around the world. “That’s really the most impressive part of this paper, I think, the wide range and the individual lineages I was able to get. That’s the part I’m most proud of,” Gimmel admits.

Gimmel collecting in the field, in California at left (photo by Chris W. Wirth), and in the Dominican Republic at right (photo by Martin Fikacek)
Sometimes collecting specimens to sample meant traveling at his own expense. For example, he took a trip to the Dominican Republic, where he beat trees and bushes, sucked up falling insects, and despaired at not having encountered the species he was looking for…until he got home, sorted through his new samples, and discovered he’d collected several of the tiny beetles without spotting them in the field.
The effort of collecting was just the beginning. Beetles in hand, Gimmel proceeded to extract DNA. While he had sequences from more than 100 species from earlier projects by his coauthors, he was adding several new genes and many more species to achieve the higher-level analysis. “Different genes evolve at different rates,” Gimmel explains. “If you use just one gene to try to determine the family tree relationships in a group, you may not have the full story.” Some genes evolve so quickly that their development is hard to trace. Some change so little over time—because the roles they play are so fundamental to the basic functions of life—that they are only helpful to determine very high-level relationships. So Gimmel and his coauthors tried to look at genes that evolve at a moderate rate, which would be more likely to reveal the relationships they were looking for.
DNA—as we all know—is microscopic. Working with it is a fiddly task. Collecting microscopic DNA from a microscopic beetle is fiddly to the max. In addition to the challenge of size, there’s another problem. “Beetles are dirty,” Gimmel explains. “They’re dirty on the outside, and their GI tract can be full of stuff.” If you don’t take your sample from a clean area, there’s a danger of accidentally contaminating it with the genes of another species, especially if this is a beetle that eats something genetically similar to itself. Fortunately, “if you can find a nice clump of good muscle tissue, you can be pretty sure that it’s clean.” (Bet you never thought of a beetle as muscular before.) So Gimmel targeted a specific area, recommended by Dr. Bocakova based on her experience with genetic sampling.
Some beetles are too small for this targeted sampling procedure. “In those cases, I would extract from the entire beetle. That means just putting it in a bath of solution and incubating it. There’s a final extraction step where you can remove the beetle body, and the DNA has already been cleaned and filtered through a column. So you can save that beetle husk [that remains].”

Tiny remnants of an undescribed species of Rentonium beetle sampled by Gimmel for the project. The black scale bars are 1 millimeter long. Photos by Lucie Gimmel
Although we have a lab suitable for extracting and amplifying DNA here at the Museum, Gimmel conducted all this lab work at Palacky University in the Czech Republic, because this project began years before he took a job here. Gimmel had visited SBMNH (in 2007) and knew our then-Curator of Entomology Michael Caterino, Ph.D. Yet he had no idea his future would lie here in Santa Barbara. “I used specimens from this institution before I knew there was going to be a job available here,” he recalls.
Having extracted the samples, Gimmel had to sort the wheat from the chaff to prepare them for sequencing. Raw DNA contains the whole mess of a specimen’s DNA, including DNA from its mitochondria. (Mitochondria are helpful structures that live within an organism’s cells and house their own DNA.) Gimmel was only interested in examining those four specially chosen genes. To sort out the mess of raw DNA, he had to amplify the genes he and his coauthors wanted to target. This meant making millions of copies of those target genes until the samples were almost entirely made up of copies of the target genes, and the rest of the DNA was proportionally insignificant.
How do you make those millions of copies? Let chain reactions do the work. You add some special ingredients (described below) to your sample. Then you heat the DNA so that the double-stranded structure unzips, leaving exposed bases. (Bases are the compounds that encode the information within DNA. They form complementary pairs: adenine/thymine and cytosine/guanine.) Then you cool the unzipped DNA down a little. This allows specialized short sequences of DNA known as primers to get to work. Assisted by a special enzyme that builds DNA, different primers will help you copy different target genes. The primers help to start the process of rebuilding only the target genes on the unzipped DNA, and the enzymes attach and draw on raw material (loose bases) you provide. The enzymes match appropriate bases to the exposed bases in the unzipped half-strands. So one original strand, unzipped, becomes two half-strands, and the primers and enzymes rebuild your target genes into two full strands. Heat, cool, and repeat; your target genes will replicate exponentially. Boom: from a whole-enchilada mess of DNA in low concentration to a nearly pure sample of your target. It sounds easy, but it took about a year to amplify all those samples.

Congratulations for making it this far. Here’s a beautiful trogossitid (Phanodesta tepaki ) to reward you. Photo by Lucie Gimmel
Pretty clever, right? But “the really clever part is the sequencing reactions,” says Gimmel. Sequencing means determining the sequence of bases in the gene, and this involves doing the same kind of process as amplification, but this time, when you provide those loose bases for use in rebuilding, some of those bases are special. They are specially dyed to make them easily identifiable (so you can spot a guanine base from a mile away, so to speak). They are also unusual in that—once used—they will stop the DNA-rebuilding reaction of a particular strand in its tracks. As soon as one is added to a strand, that strand ends. The result of running these reactions with these dyed bases in the mix is a massive pool of strands, all of which start at the beginning of the gene, but end at different lengths, depending on when the special base was attached. So some are only a single base pair long, while others may have about a thousand base pairs (depending on the length of the segment you’re sequencing). Crucially, because they’re all differently truncated copies of the same gene, they’re all following the same sequence. Since each one of those strands ends with a very visible indicator of what base is at that ending position, you only need to line them up according to length and look at the dyed bases to see the sequence. (If you’re not doing a happy dance and thinking “science is magic” right now, try re-reading the above paragraph. If molecular biology just isn’t your thing, don’t worry, we get back to beetles soon.)
For that sequencing step of the process, Gimmel delivered the samples to lab techs, who would use Sanger sequencing (that process of lining up the strands and reading the dyed bases) to turn inscrutable matter in a vial into data that could be organized by computer programs and understood by scientists. It was a hell of a lot of data, too. For every species—and remember, Gimmel was working with 395 species—the sequencers delivered information on 4,000 base pairs. Now that he had big data, Gimmel had to source the computational power to run it through programs that would analyze it. Using an external server—and drawing on his collaborators for help using the necessary programs—he ran the data through two different kinds of analyses. Because of the volume of data involved, these tasks took days to execute, even by supercomputers.
The two analyses Gimmel used are common statistical methods taxonomists use to interpret evolutionary relationships from DNA: Bayesian analysis and maximum likelihood (ML) analysis. While the simplest methods of comparing genes just look at what base pairs different samples have in common, these analyses go much further. “These methods have models of evolution built into them,” Gimmel explains. The models are grounded in observations of how certain bases are likely to be swapped for others over time, as mutations arise and spread throughout a population. Gimmel says that while ML analysis is faster, Bayesian is widely considered to be more accurate, so he wanted to run both. In the end, both analyses largely agreed on how different species were likely to be related, differing mostly in how confident they were about those relationships. Gimmel then had to take this information about likely relationships and turn it into a graphic tree that human eyes could make sense of.
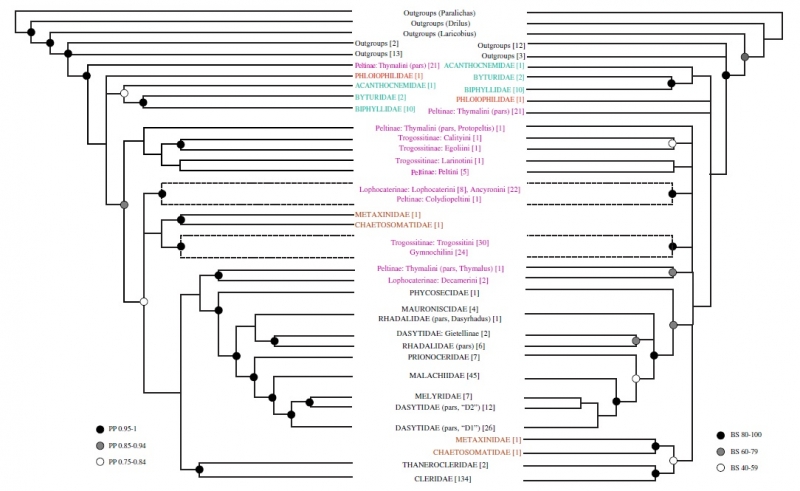
Comparison of the Bayesian and ML analyses at the higher levels of the tree. This figure stops at the tribe level (one of several intermediate levels between family and genus), but the complete tree includes limbs for each of the 395 species sampled.
That tree—which spills across six pages of the 32-page paper—bore significant fruit. The new genes Gimmel sequenced largely confirmed the previous molecular studies of his coauthors on the Melyridae and Cleridae families. However, as it struck out into new territory, the proposed tree shook up assumptions about previously understudied parts of the Cleroidea superfamily. The beetles that—in larval form on that New Zealand log—had so perplexed Gimmel were booted out of the Trogossitidae family and given their own private family (Rentoniidae, which took the root of its name from the genus, and its suffix from family naming conventions). Several other groups were similarly elevated to family level, including some reinstated families. (These had been proposed by earlier entomologists but dismissed by the wider field. If any of those scholars are alive, they must be feeling vindicated today.) Other groups merged and diverged in new ways on the basis of the analysis.
Gimmel is quick to point out that he’s not one to rock the boat for its own sake. When there were interpretive decisions to be made with ambiguous evidence, he tried to err on the side of preserving the current understanding. Yet he’s rightly proud of the results, and delighted to be receiving messages from other museum curators notifying him that they’re reorganizing their collections based on the new tree. “This study gave us a much better idea of what the groups are,” he acknowledges. He followed up on the DNA analysis by adapting a key—a tool taxonomists use to identify organisms based on their visible characteristics—to work with the new classifications.

Beetles in our collection, organized by genetic relationships. The orange cards flag families, beneath which white archival boxes are clustered by genus. Every individual in a given box belongs to the same species.
While there’s some tension between the taxonomists who rely heavily on the analysis of DNA and those traditionalists who prefer looking at an organism’s morphology, a strong family tree should resonate with both of these groups. Gimmel pointed out how the exercise of reworking the key based on the new tree required him to look differently at each species, re-evaluating what physical characteristics revealed important relationships. So learning about shared DNA can illuminate the physical organism, too, as anyone who’s scanned a human face in search of family resemblance knows. Gimmel hopes scholars testing the new key based on the new tree will identify new characteristics they’ve never noticed before, because they haven’t previously had reasons to see these particular resemblances.
Of course—because with beetles, “the fun is endless”—the work of fully understanding the superfamily Cleroidea is far from over. “In a deep evolutionary sense, we need to throw more firepower at the problem,” says Gimmel, who is leaving that task to others for the moment. His hands are full with current projects, and he won’t be putting any more metaphorical beetles in his mouth anytime soon. “I’m happy for other people to look at this problem and corroborate or refute my ideas. If there were brand new lines of data coming in, that would excite me…but I don’t want to do it all myself.”

Gimmel has to balance research with curatorial duties, like ensuring every specimen in the collection has been identified and is listed in our databases. This translates to a lot of time spent at the microscope.
For now, Gimmel is concentrating on some extremely tiny and understudied local beetles in the family Melyridae. “When people learn that I work on beetles, they want to talk to me about those big ones with the horns, the popular ones, basically. I approach it differently.” Gimmel is a beetle hipster with a purpose; he wants to help fill the gaps in our knowledge. “It’s a little bit embarrassing that we live here on Earth, and there are all these organisms flitting about, around us, and we don’t know half of them, or more.”
Mere embarrassment can turn into disaster when economic or environmental events suddenly reveal the importance of a species we can’t recognize, whose habits we don’t understand. In the early 2000s, people started to realize trees in California—including species we love and rely on for food—were being killed by beetles we couldn’t pin down to species. It’s taken years for research to catch up to these beetles, revealing that they actually belong to four different species of Shothole Borers that spread tree-killing fungus. Because we didn’t know much about these species in advance of realizing they were significant, “we lost a lot of time,” Gimmel sighs. “Now we can start to figure out, OK, what can be the potential natural enemies of these things? Do they differ in the types of lethal fungus that they’re carrying around? You couldn’t even ask these questions before, when it was just this mass of unknown beetles. If we had studied these organisms beforehand, we would have already known this. But we didn’t.” Put this way, the importance of taxonomy—the first step to understanding life—is clear. We should know our neighbors, because we never know when they’ll be significant to us.

This discreetly-placed bottle trap on our Mission Creek campus contains a pheromone lure that would attract nearby beetles in the Shothole Borer species complex. Graduate student Shelley Bennett from UCSB is monitoring our area, but the local outbreak has yet to reach the Museum’s grounds.
The magnitude of this task is astronomical. Conservative estimates of the number of undescribed insect species worldwide put this figure at about two million (other estimates range as high as 30 million). And it’s important not only to recognize different species, but to understand their family trees, because knowing those relationships can give you predictive power about how they live their lives and the roles they play in ecosystems. So it’s a good thing dedicated entomologists like Gimmel are relentless in their drive to identify and understand these animals.

Tiny beetles, big questions.
“It’s sort of like a constant itch to be scratched,” he says.
“Would you say that you’re a completionist?” I ask. (This is the kind of personality that has to score every achievement in a game or read every book in a series.)
Gimmel is thoughtful. “I have those tendencies, sure.” After a pause: “I’m trying to think of ways that manifests itself.”
“I think it’s pretty clear,” I laugh.
“Well, yeah,” he shrugs. “The sort of thing that motivates me is producing these works that illuminate all of this diversity, and exposing it to the world so it’s accessible for people to learn even more about what these beetles are doing out there. It’s just not possible to take that step without this first step, the taxonomy.”
He’s right: you can’t protect—or defend yourself from—something you don’t understand. And good luck understanding something you can’t even put a name to. Behind every charismatic conservationist stands a compulsive taxonomist, by whose obsession we have all profited.
- - -
*Yes, I know beetles are not true bugs.
†Gimmel et al. (2019) “Comprehensive phylogeny of the Cleroidea (Coleoptera: Cucujiforma).” Systematic Entomology (2019), DOI: 10.1111/syen.12338, pp. 1-31.







3 Comments
Post a CommentBeautifully written! This is a wonderful tribute to Matt and the work he is doing. I have never read such a clear and complete description of gene sequencing.
Thank you, Matt and Owen-- Your science and craft are enriching our lives!
Great article, Owen!